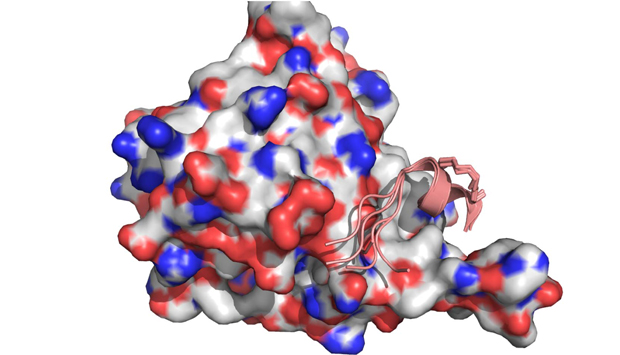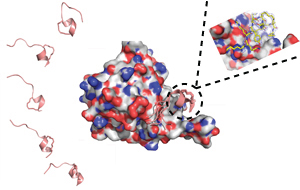
The key steps in designing stapled peptides: modeling of peptides when unbound (left), simulation of the peptide in complex with eIF4E to rationally design improved staples (center), and experimental determination of the structure of the complex to validate the method (right).
© 2014 A*STAR p53 Laboratory
The synthesis of proteins by translation of RNA relies on a protein complex called eIF4F. As formation of this complex is frequently unregulated in cancers, the ability to restore its regulation could pave the way to develop new cancer therapies. A Singapore-based team headed by Christopher Brown and David Lane from the A*STAR p53 Laboratory and Dilraj Lama at the A*STAR Bioinformatics Institute have used computer modeling to design molecules that may be able to do just that.
Regulation of eIF4F involves disrupting an interaction between two proteins in the complex, eIF4E and eIF4G. Brown and colleagues set out to mimic this disruption with artificial peptides, or small protein fragments, that bind to eIF4E in the same way as eIF4G. To do this, they controlled the peptide structures using an approach called peptide stapling.
In the context of a whole protein, peptides usually form sections of localized structure, but when these peptides are removed from a protein they become linear and highly flexible, explains Brown. “A peptide staple is a chemical modification that can link two points of a peptide together and reintroduce the structure.”
The research team designed different stapled peptides and ran computer simulations to reveal their structures, both when bound to eIF4E and when free in solution. They also synthesized the peptides and experimentally characterized their structures (see image).
Combining the information obtained by both experimentation and simulation showed where there was potential for improvement. “This meant we could design new stapled peptides, which could then be further characterized through experiments and also through computer simulations,” says Brown. “The new designs altered individual amino acids. The aim of these alterations was to either optimize the peptide when bound to eIF4E or, when in solution, to make it look more like the bound structure.”
Using the optimization process, the team produced two stapled peptides that interacted strongly with eIF4E. This required modification so that their structures would stay the same in solution as when bound to eIF4E.
“We developed a process to design highly potent inhibitors of an interaction relevant to disease,” says Brown. “Our next step is to further understand how and why certain stapled peptides are biologically active whilst others are not.”
Although these stapled peptides are not yet ready for use in treatment, Brown hopes this will change soon. “In the future, we envision developing the eIF4E stapled interacting peptides into biologically active molecules and testing them in relevant disease models.”
The A*STAR-affiliated researchers contributing to this research are from the p53 Laboratory and the Bioinformatics Institute.