Ever since penicillin became available for clinical use in the 1940s, antibiotics have saved the lives of millions of people around the globe who would otherwise have died from bacterial infections. At the same time, however, the use of antibiotics kills off all but a small number of drug-resistant bacteria, which can lead to the evolution of highly resistant bacterial strains for which antibiotics become useless. Health experts are therefore increasingly concerned about the possible emergence of a ‘super bug’ with resistance to all known drugs.
The problem may lie in the traditional methods of antibiotic development, which target bacteria’s abilities to synthesize cell walls or proteins. When an antibiotic-resistant bacterium emerges, pharmaceutical companies tend to modify the same molecule to make it re-effective against the new entity. “But soon, bacterial resistance emerges again, so this may not be a highly effective strategy to fight resistance,” says Uttam Surana, a professor at the A*STAR Institute of Molecular and Cell Biology (IMCB). “There has been a long debate about the fact that we don’t have a sufficient array of novel antibacterial compounds. So what we need are novel cellular targets.”
Cell cycles
Robert Robinson (right), Uttam Surana (center) and David Popp (left) at the IMCB
Surana specializes in investigating the division cycle of eukaryotic cells. After a cell grows for a certain period of time, chromosomes get replicated, each copy segregates away from the other, and then the cell divides into two daughter cells. The execution and coordination of the division cycle is a complex process, and any deregulation or misguiding of the cell cycle can directly endanger the stability of chromosomes and can turn normal cells into abnormal ones, such as cancer cells.
Uttam’s team uses yeast as a model system because many of the essential pathways in the cell division machinery are evolutionarily conserved from yeast to humans. “We began to think about whether lessons we learn from yeast can be utilized to identify new types of anti-proliferative compounds,” Surana says.
His idea was brought into action in 2010 in the form of collaboration with Robert Robinson, a professor and structural biologist at the IMCB, and David Popp, a research scientist in Robinson’s lab. Robinson and Popp are interested in the functions of bacterial actins and tubulins—cytoskeletal elements that form filaments. “These are essential proteins for cell division and normal growth. Interfering with them is an attractive strategy for the development of novel antibiotics,” says Robinson.
Bacterial filaments
Common to bacterial cells and the eukaryotic cells that constitute our body are proteins called actins and tubulins, which serve vital roles in the cell cycle. In bacterial cells, actins generally either form part of the machinery that segregates plasmids—genetic fragments capable of autonomous replication—or act as cell-shape determining factors required for normal cell growth. In eukaryotic cells, tubulin is involved in the formation of microtubules or centrosomes, both of which have key roles in the progression of the cell cycle. The researchers at the IMCB are particularly interested in the bacterial equivalent of tubulin, known as FtsZ. This protein forms a ring that is attached to the cell membrane, where it participates in the formation of two daughter cells.
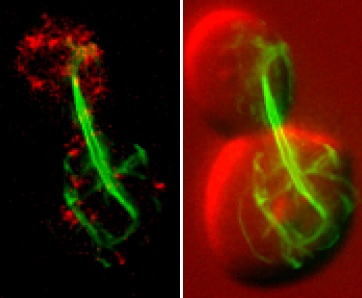
The budding yeast Saccharomyces cerevisiae tagged using green fluorescent protein to highlight a bacterial division protein that assembles into large polymers
When put under the control of inducible yeast promoter DNA, bacterial actin or FtsZ can be expressed in yeast grown in an appropriate medium. The expressed actin or tubulin begins to form filaments and interfere with the functioning of the yeast’s cell division machinery. Consequently, cells stop growing and eventually die. This phenomenon can be used to design effective screening strategies for the identification of novel antibacterial agents. The researchers have collected about 30 cytoskeleton proteins from various bacteria, both pathogenic and non-pathogenic, including the common pathogen Escherichia coli.
One of the most interesting observations was that the tubulin-like bacterial FtsZ plays an analogous role to eukaryotic actin, whereas bacterial actins segregate DNA in a manner similar to microtubules in eukaryotic cells. “The proteins appear to have swapped roles,” Robinson says. Popp found FtsZ to be reasonably similar across many bacteria, whereas bacterial actin comes in a variety of filament geometries. “This gives us an opportunity to search for a pan-bacteria antibiotic targeted at FtsZ or to search for an antibiotic directed against a specific species or cluster of species through targeting bacterial actins. As bacterial actins and FtsZ filaments are very different from human actin and tubulin filaments, disruptive agents are unlikely to cross-react,” comments Robinson.
The researchers are now looking to find compounds that can prevent the polymerization of bacterial actins or tubulins in yeast because such agents could allow yeast cells to grow. “This strategy permits us to select promising antibacterial compounds that are nontoxic to eukaryotic cells.”
Communications among bacteria

Lianhui Zhang from the IMCB
Lianhui Zhang, a professor at the IMCB, also shares the concerns about the growing problem of drug resistance. “Bacteria produce lots of biochemical weapons such as toxins and enzymes that can interfere with our immune systems,” Zhang explains. “It would be great if we can prevent bacterial cells from producing such biochemical weapons. Without them, bacteria cannot damage our immune systems.”
Bacteria are single-cell organisms, but they are not acting individually in their community. Instead, they communicate with each other using chemical signaling molecules. According to Zhang, many bacteria synchronize gene expression among family members and coordinate a range of important biological functions, such as virulence and biofilm formation. Zhang is one of the pioneers who discovered that such a cell-to-cell communication mechanism, known as ‘quorum sensing’, is widely conserved among various bacteria.
A hint from rotten cabbage
Since joining the A*STAR in 1998, Zhang’s first objective has been to test the idea that quorum sensing could represent a target for the control of bacterial infections. He focused on the quorum sensing signals and in 2000 reported an enzyme that inactivates N-acyl homoserine lactones (AHLs), one of the most widely conserved quorum-sensing signals. When looking for a model bacterial pathogen to test his idea, Zhang found a Chinese cabbage in his kitchen that had gone rotten overnight. Following his instincts, he isolated the bacterium that caused the damage to the vegetable, and in 2001 his team demonstrated that it was feasible and practical to control bacterial infections by using the AHL-inactivation enzyme to quench bacterial quorum sensing1. He later identified another enzyme associated with the degradation of AHL signals and several regulatory proteins in the AHL signaling pathways.
Further studies
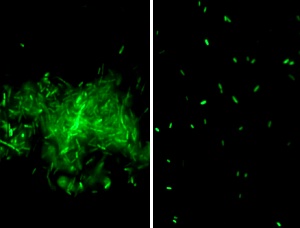
DSF induces bacterial biofilm dispersion. (Left) X. campestris biofilm. (Right) Disperse biofilm after addition of DSF.
After demonstrating the feasibility of using the quorum-sensing molecule as a target for the control of bacterial infections, Zhang’s team moved on to identify and characterize more quorum sensing signals. “We realized that AHL molecules might control only one portion of pathogenicity in some bacterial pathogens and not all bacteria produce AHLs,” Zhang says. His team became the first to identify the chemical structure of an important molecule called diffusible signal factor (DSF) in Xanthomonas campestris, a bacterium that causes plant diseases. The researchers are also studying how pervasively DSF is conserved among bacterial species, and through that study have identified a few analogues of DSF in a range of microorganisms including human bacterial pathogens. Their findings indicate that similar to AHL, DSF represents a new family of widely conserved quorum sensing signals in Gram-negative bacteria.
The team has begun searching for chemicals capable of controlling quorum sensing, but Zhang indicates there are still plenty of things to investigate before therapeutic applications become possible. “Bacterial infections commonly involve a few hundred genes, most of which are regulated by various signaling mechanisms. Our goal is to develop a drug or drug combination to silence these genes,” Zhang says. “But first, I will try to identify more chemical signals and signaling mechanisms because each system controls a subset of genes. When we have a fair understanding of these signal pathways, we will have better ideas on how to block cell-cell communication and control bacterial infections.”



